Should I install PyCytoData?
In case you haven’t heard, PyCytoData provides a unified
framework for analyzing CyTOF data in python. The supported workflow includes File IO, preprocessing, and
dimension reduction and visualization via CytofDR. While we believe in the future of PyCytoData, we recognize
that many people prefer a leaner package without the baggage of an ecosystem. Thus, we kindly made PyCytoData
to only those who need it. In this tutorial, we walk you through whether you need to install PyCytoData
and help you make the best decisions.
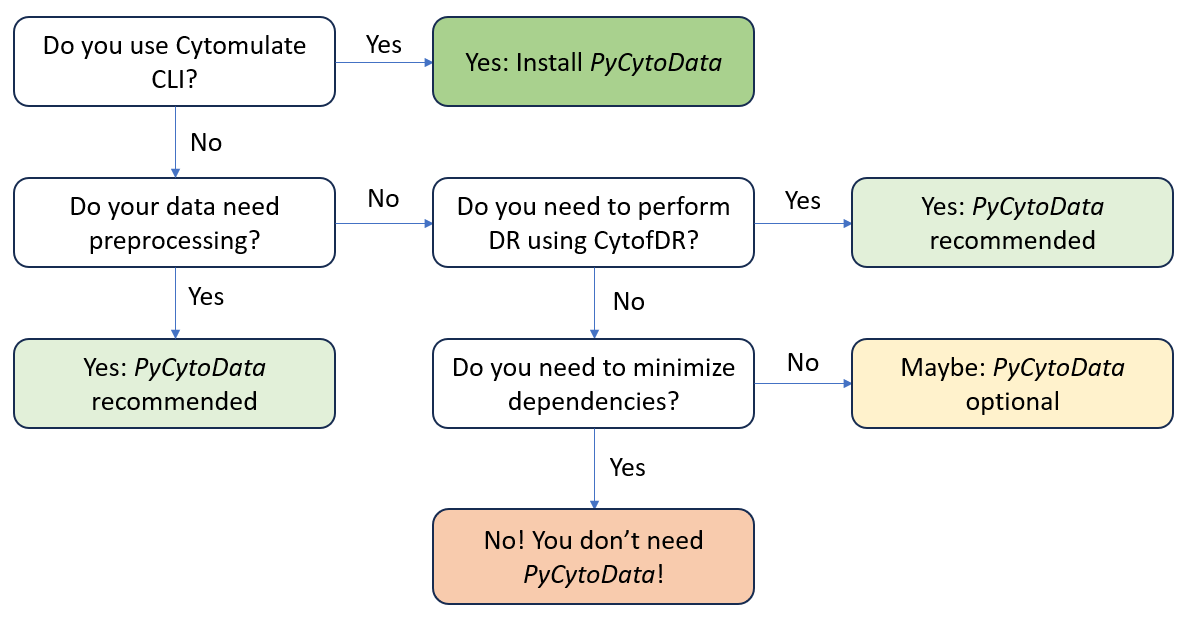
Decision at a Glance
If you need a TLDR, we create a nifty flowchart for you to follow. At each step, ask yourself the question
and you will arrive at a conclusion. We want to emphasize that currently only the Cytomulate CLI strictly
requires PyCytoData because we don’t want to maintain two branches of the same IO code. For all other
workflows, you can judge the necessity of Cytomulate yourself.
How is PyCytoData optional?
Currently, we need PyCytoData for two functionalities:
The Cytomulate CLI
Outputting to a PyCytoData object
For the former, we require PyCytoData because we rely on its File IO operations so that we don’t have
reinvent a great module. For the latter, users have the option to use the sample_to_pycytodata method
or just the regular sample method. In the case that PyCytoData is not installed, user can have
NumPy arrays returned instead. As you may see, we do not rely on PyCytoData for the core model and
estimation procedures. PyCytoData is rather just a convenience feature for those who need it.
In terms of implementation, we check whether PyCytoData is importable at the beginning. If not, no
ImportError is raised until absolutely necessary. This means that unless users explicitly call
smaple_to_pycytodata without a proper installation, we do not complain! Of course, this implementation
has its downsides: namely, it can surprise those who are not aware of the optional dependency. We will
revisit this in the future.
What are added benefits of PyCytoData?
The main reason to use PyCytoData is its convenience. Specifically, it can handle batches, metadata,
and working with expression matrices easily. For those who want a wrapper around arrays, we believe that
PyCytoData is a good fit. Of course, let’s not forget the File IO capabilities and downstream integration
with CytofDR.
On the note of downstream analyses, PyCytoData is being actively developed. So, there will be some neat
features coming in the futrue when we add more tools and integrate more libraries.
Should I skip PyCytoData for CytofDR directly?
If you are asking this question, then YES! The CytofDR package provides a more complete set of
tools for benchmarking and performing DR in CyTOF. While PyCytoData can have workarounds to allow
these functionalities, we recommend using CytofDR directly. However, you are always welcomed to
install PyCytoData to manage other aspects of the CyTOF workflow.